Research
Bacteria as single living organisms are exposed to rapidly changing external conditions. Given strong selection pressure, it is not surprising that bacteria have evolved sophisticated signaling systems to constantly monitor changes in external parameters, such as acidity, nutrients, eukaryotic signals and to adapt their structure, physiology and behavior accordingly.
Research in Kirsten Jung's lab focuses on elucidating the molecular mechanisms of stimulus perception under acid stress in Escherichia coli and other γ-proteobacteria (Area 1). We study the uptake, excretion and homeostasis of primary metabolites, such as pyruvate, in Escherichia, Salmonella and Vibrio (Area 2). We elucidate the role of RNA modifications during abiotic and biotic stress response in Escherichia and Vibrio (Area 3). We also investigate translation elongation factor P (EF-P), a protein that alleviates ribosome stalling at polyproline stretches, and its post-translational modification systems (Area 4). Last but not least, we are working on identifying new intracellular targets of natural compounds (Area 5).
Research Area 1: Acid stress sensing in Escherichia coli and other γ-proteobacteria
Acid resistance is an important property of many bacteria to survive in acidic environments like the human gastrointestinal tract. E. coli, for example, has several sophisticated signaling systems to sense and appropriately respond to environmental acid stress by regulating the activity of five inducible acid resistance systems. We investigate how Gram-negative bacteria sense environmental acidity using membrane-integrated and cytosolic pH sensors.
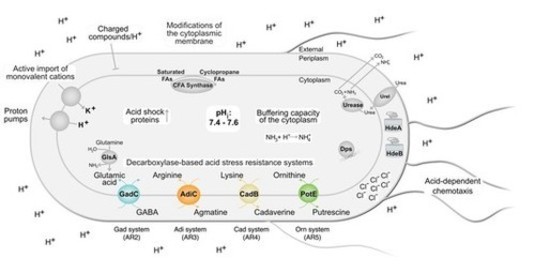
Figure 1: Overview of mechanisms in Gram-negative proteobacteria to cope with acidic environments (for details, Schwarz, J., Schumacher, K., Brameyer, S., Jung, K. (2022) Battle of bacteria against acidity. FEMS Microbiol. Rev., DOI: 10.1093/femsre/fuac037).
Selected Publications:
- Schumacher, K., Gelhausen, R., Backofen, R., Kion-Crosby, W., Barquist, L., Jung, K. (2023) Ribosome profiling reveals the fine-tuned response of Escherichia coli to acid stress. mSystems, Nov 1:e0103723. doi: 10.1128/msystems.01037-23.
- Schwarz, J., Brameyer, S., Hoyer, E., Jung, K. (2023) The interplay of AphB and CadC to activate acid resistance of Vibrio campbellii. J. Bacteriol., 205: e0045722. doi: 10.1128/jb.00457-22.
- Schumacher, K., Brameyer, S., Jung, K. (2023) Bacterial acid stress response: from cellular changes to antibiotic tolerance and phenotypic heterogeneity. Curr. Opin. Microbiol., 75: 102367. https://doi.org/10.1016/j.mib.2023.102367.
- Brameyer, S., Schumacher, K., Kuppermann, S., Jung, K. (2022) Division of labor and collective functionality in Escherichia coli under acid stress. Commun. Biol., 5: 327.
- Martini, L., Brameyer, S., Hoyer. E., Jung. K.*, Gerland, U.* (2021) Dynamics of chromosomal target search by a membrane-integrated one-component receptor. PloS Comp. Biol., 17: e1008680.
- Brameyer, S., Hoyer, E., Bibinger, S., Burdack, K., Lassak, J., Jung, K. (2020) Molecular design of a signaling system influences noise in protein abundance under acid stress in different gamma-proteobacteria. J Bacteriol, 202 (16): e00121-20.
- Brameyer, S., Rösch, T.C., Andarib, J.A., Hoyer, E., Schwarz, J., Graumann, P.L., Jung, K. (2019) DNA-binding directs the localization of a membrane-integrated receptor of the ToxR family. Commun. Biol., 2: 4.
- Fritz, G., Koller, C., Tetsch, L., Haneburger, I., Burdack, K., Jung, K., Gerland, U. (2009) Induction kinetics and feedback inhibition of a conditional stress response system in Escherichia coli, J. Mol. Biol. 393: 272–286.top
Research Area 2: Molecular mechanisms of stimulus perception by histidine kinase/response regulator systems and regulation of uptake and excretion of primary metabolites
Bacteria sense and respond to various stress conditions by employing so called two-component systems. These systems consist of a histidine kinase and a response regulator, which sense environmental stimuli, transduce information via phosphorylation and induce a cellular response. Escherichia coli, for example, contains 32 of these systems.
We are studying how sensing and uptake of the primary metabolite pyruvate is coordinated by the LytS/LytTR-type histidine kinase/response regulator system BtsS/BtsR and the transporter BtsT in Escherichia, Vibrio and Salmonella species. Currently, our studies focus on the indentification of the ligand sensed by the second LytS/LytTR-type histidine kinase/response regulator system in E. coli, the YpdA/YpdB system and the function of target gene product YhjX.
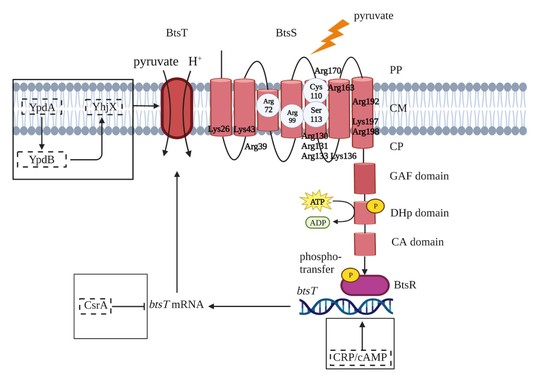
Figure 2: Schematics of the pyruvate-responsive LytS/LytTR-type histidine kinase/response regulator BtsS/BtsR system of E. coli. Extracellular pyruvate stimulates the BtsS/BtsR signaling cascade that regulates the expression of btsT, which encodes a high-affinity pyruvate transporter. The system is embedded in a complex regulatory network (proteins with regulatory function are shown in dashed boxes). BtsS is a receptor with seven transmembrane helices. The position of all lysines and arginines in the transmembrane domain is marked. Amino acids involved in pyruvate binding, Arg72, Arg99, Cys110, and Ser113, are labeled in a gray circle. See Qiu et al., 2023 for details.
Selected Publications:
- Qiu, J., Gasperotti, A., Sisattana, N., Zacharias, M., Jung, K. (2023) The LytS-type histidine kinase BtsS is a 7-transmembrane receptor that binds pyruvate. mBio, Sep 1:e0108923. DOI: 10.1128/mbio.01089-23.
- Paulini, S., Fabiani, F.D., Weiß, A.S., Moldoveanu, A.L., Helaine, S., Stecher, B., Jung, K. (2022) The biological significance of pyruvate sensing and uptake in Salmonella enterica Serovar Typhimurium. Microorganisms, 10: 1751
- Göing, S., Gasperotti, A.F., Yang, Q., Defoirdt, T., Jung, K. (2021) Insights into a pyruvate sensing and uptake system in Vibrio campbellii and its importance for virulence. J. Bacteriol. 203: e00296-21
- Steiner, B.D., Eberly, A.R., Hurst, M.N., Zhang, E., Behr, S., Jung, K., Hadjifrangiskou, M. (2018) Evidence of cross-regulation in two closely-related pyruvate-sensing systems in uropathogenic Escherichia coli, J. Membr. Biol., 251(1):65-74.
- Vilhena, C., Kaganovitch, E., Shin, J.Y., Grünberger, A., Behr, S., Kristoficova, I., Brameyer, S., Kohlheyer, D., Jung, K. (2018) A single cell view of the BtsSR/YpdAB pyruvate sensing network in Escherichia coli and its biological relevance, J. Bacteriol., 200: e00536-17.
- Behr, S., Kristoficova, I., Wittig, M., Breland, E.J., Eberly, A.R., Sachs, C., Schmitt-Kopplin, P., Hadjifrangiskou, M., Jung, K. (2017) Identification of a high-affinity pyruvate receptor in Escherichia coli, Sci. Rep., 7: 1388.
- Fried, L., Behr, S., Jung, K. (2013) Identification of a target gene and activating stimulus for the YpdA/YpdB histidine kinase/response regulator system in Escherichia coli, J. Bacteriol. 195: 807-815
- Kraxenberger, T., Fried, L., Behr, S., Jung, K. (2012) First insights into the unexplored two-component system YehU/YehT in Escherichia coli, J. Bacteriol. 194: 4272-4284.top
Research Area 3: The role of m6A-mRNA modification to control stress response and phage replication
In this project we are studying the role of mRNA modifications to control stress response in E. coli and phage replication in the marine bacterium Vibrio campbellii ATCC BAA-116, a model organism for quorum sensing.
Figure 3: The role of mRNA modification in bacteria
Selected Publications:
- Petrov, D.P., Kaiser, S., Kaiser, S.*, Jung, K.* (2022) Opportunities and challenges to profile mRNA modifications in Escherichia coli. Chembiochem. 23: https://doi.org/10.1002/cbic.202200270.
- Reichle, V., Petrov, D., Weber, V., Jung, K., Kellner, S. (2019) NAIL-MS reveals the repair of 2-methylthiocytidine by AlkB in E. coli. Nat. Commun., 10: 5600.
Research Area 4: Post-translational modifications of elongation factor P (EF-P) and the role of poly-proline motifs
The fundamental process of protein synthesis is catalyzed by ribosomes. We have recently demonstrated that bacterial ribosomes become arrested when translating proteins contain consecutive polyproline stretches, and that this arrest is alleviated by the translation elongation factor EF-P. Furthermore, we showed that the post-translational β-lysinylation of lysine34 of Escherichia coli EF-P by the enzymes EpmA and EpmB and the rhamnosylation of arginine32 of Shewanella EF-P by EarP, respectively, are critical for the rescue activity of EF-P. EF-P is aminopentanoylated in Bacillus and other members of the Firmicutes. In contrast, the EF-Ps of Actinobacteria – specifically, Clostridium glutamicum, Streptomyces coelicolor and Mycobacterium tuberculosis – alleviate ribosome stalling at polyproline motifs without the need for any activating post-translational modification. In our recent study we decrypted the functional design of unmodified translation elongation factor P in E. coli.
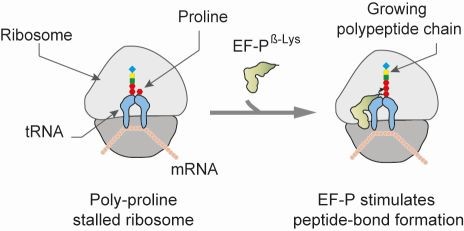
Figure 4: EF-P prevents ribosome stalling at polyproline stretches.
Many virulence factors contain polyproline stretches, which explains why modified EF-P is critical for bacterial pathogenicity. Polyproline motifs are required for the regulation of copy number or dimerization or catalytic activity of certain proteins. The project focuses on the elucidation and characterization of synthetic and novel post-translational modification pathways of EF-P.
Selected Publications:
- Tomasiunaite, U., Kielkowski, P., Krafczyk, R., Forné, I., Imhof, A., Jung, K. (2024) Decrypting the functional design of unmodified translation elongation factor P. Cell Rep., published: April 16, 2024, https://doi.org/10.1016/j.celrep.2024.114063
- Pinheiro, B., Scheidler, C.M., Kielkowski, P., Schmid, M., Forné, I., Ye, S., Reiling, N., Takano, E., Imhof, A., Sieber, S. A., Schneider, S., Jung, K. (2020) Structure and function of a novel elongation factor P subfamily in Actinobacteria. Cell Rep., 30: 4332-4342.e5.
- Pfab, M., Kielkowski, P., Krafczyk, R., Volkwein, W., Sieber, S.A., Lassak, J., Jung, K. (2021) Synthetic post-translational modifications of elongation factor P using the ligase EpmA. FEBS J., 288: 663-677.
- Motz, L., Jung, K. (2018) The role of polyproline motifs in the histidine kinase EnvZ, PLoS One, 13(6): e0199782.
- Qi, F., Motz, M., Jung, K., Lassak, J. and Frishman, D. (2018) Evolutionary analysis of polyproline motifs in Escherichia coli reveals their regulatory role in translation, PLoS Comp. Biol. 14(2):e1005987.
- Starosta, A.L., Lassak, J., Atkinson, G.C., Peil, L., Woolstenhulme, C.J., Virumäe, K., Buskirk, A., Tenson, T., Remme, J., Jung, K., Wilson, D.N. (2014) A conserved proline triplet in Val-tRNA synthetase and the origin of elongation factor P, Cell Rep., 9: 476-483.
- Lassak, J., Keilhauer, E., Fürst, M., Wuichet, K., Gödeke, J., Starosta, A.L., Chen, J., Søgaard-Andersen, L., Rohr, J., Wilson, D.N., Häussler, S., Mann, M., Jung, K. (2015) Arginine-rhamnosylation as new strategy for post-translational modification of translation elongation factor P, Nat. Chem. Biol., 11: 266-70, doi:10.1038/nchembio.1751.
- Ude, S., Lassak, J., Starosta, A. L., Kraxenberger, T., Wilson, D.N., Jung, K. (2013) Translation elongation factor EF-P alleviates ribosome stalling at polyproline stretches. Science, 339: 82-85.
Research Area 5: Chemical Biology and the identification of protein targets of natural compounds
There are about 500,000 molecules in the communication between pro- and eukaryotes. We use bacterial model organisms to study the effect of natural compounds such as fimbrolides, quorum quenchers or eukaryotic hormones on the phenotypic behavior of bacteria, such as quorum sensing and chemotaxis. The project also focuses on the identification of the cellular targets of these compounds.
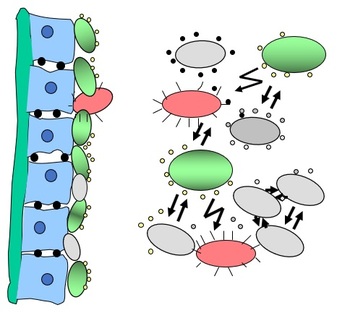
Figure 5: Chemical communication between pro- and eukaryotes.
Selected Publications:
- Mostert, D., Bubeneck, W.A., Rauh, T., Kielkowski, P., Itzen, A., Jung, K., Sieber, S.A. (2024) Pronucleotide probes reveal a diverging specificity for AMPylation vs UMPylation of human and bacterial nucleotide transferases. Biochemistry, 63:651-659. doi: http://doi.org/10.1021/acs.biochem.3c00568.
- Weigert Muñoz, A., Hoyer, E., Schumacher, K., Grognot, M., Taute, K., Hacker, S., Sieber, S.A., Jung, K. (2022) Eukaryotic catecholamine hormones influence the chemotactic control of Vibrio campbellii by binding to the coupling protein CheW. Proc. Natl. Acad. Sci. USA. 119: e2118227119. doi: https://doi.org/10.1073/pnas.2118227119.
- Rauh, T., Brameyer, S., Itzen, A., Kielkowski, P., Jung, K., Sieber, S.A. (2020) MS-based in situ proteomics reveals AMPylation of host proteins during bacterial infection, ACS Infect. Dis., 6: 3277-3289.
- Zhao, W., Lorenz, N., Jung, K., Sieber, S.A. (2016) Mechanistic analysis of aliphatic β-lactones in Vibrio harveyi reveals a quorum sensing independent mode of action, Chem. Commun., 52: 11971-11974
- Zhao, W., Lorenz, N., Jung, K., Sieber, S.A. (2016) Fimbrolide natural products disrupt bioluminescence of Vibrio harveyi by targeting autoinducer biosynthesis and luciferase activity, Angew. Chem. Int. Ed. Engl. 55(3): 1187-91.
- Rossmann, F.S., Wobser, D., Racek, T., Puchalka, J., Rabener, E.M., Reiger, M., Hendrickx, A.P.A., Diederich, A., Jung, K., Klein, C., Huebner, J. (2015) Phage-mediated dispersal of biofilm and distribution of bacterial virulence genes is induced by Quorum Sensing, PLoS Pathog., 11(2): e1004653.
- Chu, Y., Nega, M., Wölfle, M., Plener, L., Grond, S., Jung, K., Götz, F. (2013) A new class of quorum quenching molecules from Staphylococcus species affects communication of Gram-negative bacteria, PLoS Pathog., 9(9): e1003654.

